PGM Examples - PGM Class to Set Species Splits
Jump to navigation
Jump to search
Navigation: PGMs ➔ Example PGM Files ➔ Class to Set Species Splits
Related Links: Defining a Class, SplitFlow
PGM Class to Set Species Splits
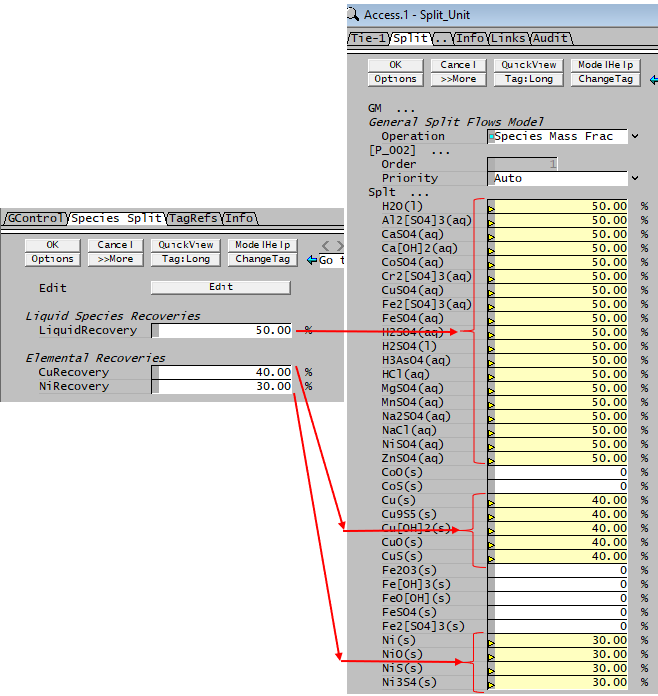
The following Class Example PGM file - Class_Species.pgm (given under the table) allows the user to set the species split in a Split - Species Mass Frac based on any of the following requirements:
| Purpose | Description | Example function call using the SpeciesClass Defined in the Class_Species.pgm |
|---|---|---|
| Set Mass Fraction Split based on One (1) given Element | Set the split Mass Fraction of all species containing a given element in a user defined phase, i.e. the user can send 80% of all of the Copper in the solid phase to a required stream. |
Init("Cu", "s") ;call initialisation function
IonSplit("Splitter.GM.IOs.[P_002]", 80) ;call the split function
|
| Set Mass Fraction Split based on Two (2) given Elements | Set the split Mass Fraction of all species containing two given elements in a user defined phase, i.e. the user can send 75% of all species that contain both Copper and Sulphur in the solid phase to a required stream. This method selects CuS(s), Cu2FeS4(s), etc., but not Cu(s) or NiS(s) - the species must contain BOTH elements. |
Init2("Cu", "S", "s") ;call initialisation function
IonSplit("Splitter.GM.IOs.[P_002]", 75) ;call the split function
|
| Set Mass Fraction Split based on One (1) given Individual Phase | Set the split of all species in an Individual Phase to a stream, i.e. send 2% of all (aq) species to a stream. |
IndPhaseSplit("Splitter.GM.IOs.[P_002]", 2, "aq")
|
| Set Mass Fraction Split based on One (1) given Phase | Set the split of all species in a Phase to a stream, i.e. send 5% of all liquid species to a stream. |
PhaseSplit("Splitter.GM.IOs.[P_002]", 5, "l")
|
| Set Mass Fraction Split based on Density | Set the split of all species in a Phase to a stream based on density separation, i.e. send 100% of all solids with density > 3000 kg/m3 to a stream. |
DensitySplit("Density_Splitter_001.GM.IOs.[P_HeavySolids]", 3000, "s")
|
PGM FILE: Class_Species.pgm
;=======================================================================================
;--- SysCAD General Controller - Species Splitter class ---
long SpeciesCount
; Find the total number of species in the project
SpeciesCount = SDB.SpeciesCount()
Class SpeciesClass
Long ReqPhase, isp, SplitPhase, NumIonSpecies
Array IonIndex
;------------------------------------------------------------------------------
; Initialisation function to set up an array with all of the species in the
; required phase (GPhase) that contain the user defined element (Ion)
Function Init(String Ion, String GPhase)
IonIndex.SetLen(25)
IonIndex.SetAll(0)
NumIonSpecies = 0
long Cmp1, SpPhase, ReqdPhase, j
; Find the index of the required phase
ReqPhase = SDB.FindPhase(GPhase)
isp = 0
j = 0
; Scroll through all the species in the required phase in the project to find
; ones that contain the required element
while (isp < SpeciesCount)
SpPhase = SDB.SpIPhaseNo(isp)
if (SpPhase == ReqPhase)
Cmp1 = SDB.SpElemMoles(isp, Ion)
if (Cmp1 > 0)
IonIndex.SetAt(j,isp)
j = j + 1
endif
endif
isp = isp + 1
endwhile
NumIonSpecies = j
return 0
EndFunct
;------------------------------------------------------------------------------
; Initialisation function to set up an array with all of the species in the
; required phase (GPhase) that contain both user defined elements (Ion1 and Ion2)
Function Init2(String Ion1, String Ion2, String GPhase)
IonIndex.SetLen(20)
IonIndex.SetAll(0)
NumIonSpecies = 0
long Cmp1, Cmp2, SpPhase, ReqdPhase, j
; Find the index of the required phase
ReqdPhase = SDB.FindPhase(GPhase)
isp = 0
j = 0
; Scroll through all the species in the required phase in the project to find
; ones that contain both the required elements
while (isp < SpeciesCount)
SpPhase = SDB.SpIPhaseNo(isp)
if (SpPhase == ReqdPhase)
Cmp1 = SDB.SpElemMoles(isp, Ion1)
Cmp2 = SDB.SpElemMoles(isp, Ion2)
if ((Cmp1 > 0) AND (Cmp2 > 0))
IonIndex.SetAt(j,isp)
j = j + 1
endif
endif
isp = isp + 1
endwhile
NumIonSpecies = j
return 0
EndFunct
;------------------------------------------------------------------------------
; Function to split a single required species to a stream at a user defined value
Function SpeciesSplit(String SplitTag, Real ReqValue, String ReqdSpecies)
String tempsplit
tempsplit = Concatenate(SplitTag, ".Splt.", ReqdSpecies ," (%)")
[tempsplit] = ReqValue
return 0
EndFunct
;------------------------------------------------------------------------------
; Function to split a single species with a required index to a stream at a user defined value
Function SpeciesIndexSplit(String SplitTag, Real ReqValue, long SpeciesIndex)
String tempsplit, RequiredSpecies
RequiredSpecies = SDB.SpSymbol(SpeciesIndex)
tempsplit = Concatenate(SplitTag, ".Splt.", RequiredSpecies ," (%)")
[tempsplit] = ReqValue
return 0
EndFunct
;------------------------------------------------------------------------------
; Function to split all species containing a required element(s) to a stream at a user defined value
; Must have initialised the array first using Init or Init2
Function IonSplit(String SplitUnit, Real SplitValue)
String TempSp
long js
js = 0
while (js < NumIonSpecies)
isp = IonIndex.GetAt(js)
TempSp = SDB.SpSymbol(isp)
SpeciesSplit(SplitUnit, SplitValue, TempSp)
js = js + 1
endwhile
return 0
EndFunct
;------------------------------------------------------------------------------
; Function to split all species in a required individual phase to a stream at a user defined value
Function IndPhaseSplit(String SplitUnit, Real SplitValue, String ReqdIPhase)
String TempSp
long SpPhase
SplitPhase = SDB.FindIPhase(ReqdIPhase)
isp = 0
while (isp < SpeciesCount)
SpPhase = SDB.SpIPhaseNo(isp)
if (SpPhase == SplitPhase)
TempSp = SDB.SpSymbol(isp)
SpeciesSplit(SplitUnit, SplitValue, TempSp)
endif
isp = isp + 1
endwhile
return 0
EndFunct
;------------------------------------------------------------------------------
; Function to split all species in a required phase to a stream at a user defined value
Function PhaseSplit(String SplitUnit, Real SplitValue, String ReqdPhase)
String TempSp
long SpPhase
SplitPhase = SDB.FindPhase(ReqdPhase)
isp = 0
while (isp < SpeciesCount)
SpPhase = SDB.SpPhaseNo(isp)
if (SpPhase == SplitPhase)
TempSp = SDB.SpSymbol(isp)
SpeciesSplit(SplitUnit, SplitValue, TempSp)
endif
isp = isp + 1
endwhile
return 0
EndFunct
;------------------------------------------------------------------------------
; Function to split all species in a stream at a user defined value, useful for initialising values after priority change.
Function SetAll(String SplitUnit, Real SplitValue)
String TempSp
isp = 0
while (isp < SpeciesCount)
TempSp = SDB.SpSymbol(isp)
SpeciesSplit(SplitUnit, SplitValue, TempSp)
isp = isp + 1
endwhile
return 0
EndFunct
;------------------------------------------------------------------------------
; Function to split all species in a required phase based on Density to a stream at a user defined value
Function DensitySplit(String SplitUnit, Real DensitySplitValue, String ReqdPhase)
String TempSp
long SpPhase
SplitPhase = SDB.FindPhase(ReqdPhase)
isp = 0
while (isp < SpeciesCount)
SpPhase = SDB.SpPhaseNo(isp)
if (SpPhase == SplitPhase)
TempSp = SDB.SpSymbol(isp)
if SDB.SpDensity(isp,298.15,101.325) > DensitySplitValue
SpeciesSplit(SplitUnit, 100, TempSp)
Else
SpeciesSplit(SplitUnit, 0, TempSp)
endif
endif
isp = isp + 1
endwhile
return 0
EndFunct
EndClass
One of the advantages of using this class when doing species splits, is that if the user adds species to the project, this class will automatically include it in the split.
For an example PGM that uses this class, see below.
Example PGM file including the Species Class
The following is an example of a PGM file that uses the above splitter class: