How to perform a Species Mass Balance
Navigation: PGMs ➔ Example PGM Files ➔ Species Mass Balance
| Simple Examples | Subroutines Examples | Dynamic Examples | Steady State Overall Mass Balance | Array and Matrix Examples | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Basic Layout | Simple Calculations | Initialise PreStart | Multi-Step Trigger | Checking Project | Counter, While and Random | Belt Filter Wash Loss | Startup Actions | Mass Balance | Mass Balance | Species Balance | Elemental Balance | Lookup Value | Set Values | Tridiagonal System |
Related Links: PGM Files using Class and Functions, TagSelect Class, FAQ (Mass & Energy Balance)
It is often important to perform species mass balance to account for the total in / out of the species and how much the species has reacted in reactions. To obtain a quick balance of a species, we can do the following:
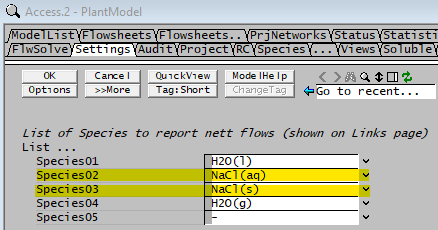
- Add the Species of interest to the PlantModel Settings Tab.
- Add a PGM file using TagSelect Class to sum up the total in, out and reacted amount and perform a balance.
Adding Species to the tracking List
We will be using NaCl as an example to for this document.
From the View - Plant Model command, go to Settings Tab, add in the species as shown below:
Use PGM file to perform mass balance
To perform a mass balance of the species, we recommend using the TagSelect Class at the end of project solve, some examples are given below:
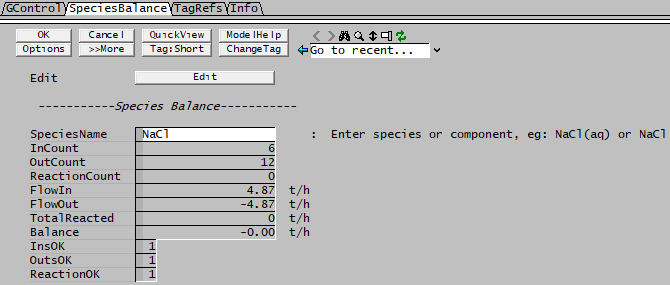
Example 1 - showing input, output and reacted amount
;--- SysCAD General Controller (PGM) program logic file ---
; Revision: 2 Date: Jan 2023 Author: SysCAD Team
;--- variable declarations ---
PageLabel "SpeciesBalance"
TextLabel , " -----------Species Balance-----------", ""
TagSelect Inputs, Outputs, Reactions
Array InputsList, OutputsList, ReactionsList
String SpeciesName*{Comment(" Enter species or component, e.g.: NaCl(aq) or NaCl ")}
integer InCount@, OutCount@, ReactionCount@
real FlowIn@("Qm","t/h"), FlowOut@("Qm","t/h"), TotalReacted@("Qm","t/h"), Balance@("Qm","t/h")
Checkbox InsOK@, OutsOK@, ReactionOK@
string flow_tag, Criteria, Criteria_Tag1, Criteria_Tag2
Sub CalcBalance()
; find all true feeders (not connected) and makeup sources
InsOK= Inputs.Exec("([UnitType]=='FeederSink' AND ([State]==1 AND [Operation]!=11)) OR ([UnitType] == 'MakeupSource')", false, false)
InCount = Inputs.GetLen()
; sum all input flows
flow_tag = Concatenate(".Links.Nett.Qm.", SpeciesName, " (t/h)")
Inputs.GetValues(flow_tag, InputsList)
FlowIn = InputsList.Sum()
; find all true sinks (not connected)
OutsOK = Outputs.Exec("(([UnitType]=='FeederSink' AND [State]==2) OR ([UnitType] == 'DiscardSink'))", false, false)
OutCount = Outputs.GetLen()
; sum all output flows
flow_tag = Concatenate(".Links.Nett.Qm.", SpeciesName, " (t/h)")
Outputs.GetValues(flow_tag, OutputsList)
FlowOut = OutputsList.Sum()
; find all user-specified reactions (excluding units with auto-generated reactions such as GFEM or TCE reactors)
ReactionOK = Reactions.Exec("[SubTypeList] like 'RB.*'", false, false)
ReactionCount = Reactions.GetLen()
; sum all Reacted flows
flow_tag = Concatenate(".Links.Nett.Qm.", SpeciesName, " (t/h)")
Reactions.GetValues(flow_tag, ReactionsList)
TotalReacted = ReactionsList.Sum()
; perform mass balance, note that flow out and reacted amount are reported as negative numbers.
Balance = FlowIn + FlowOut + TotalReacted
EndSub
Sub TerminateSolution()
;--- Logic executed after last iteration (when solver is stopped)
CalcBalance()
EndSub
$
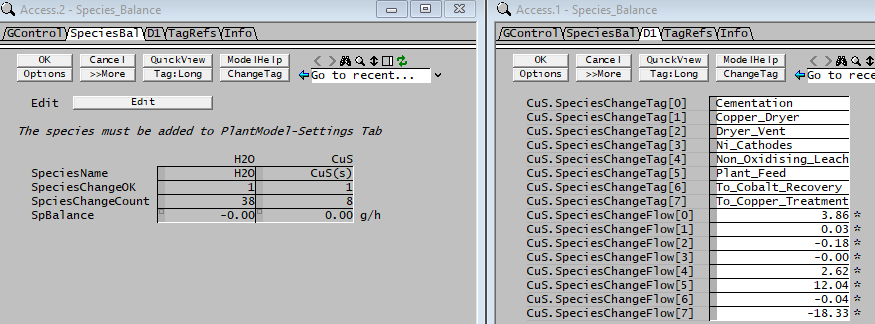
Example 2 - Showing all species flow change
Class Class_SpeciesBalance
string flow_tag, Criteria_Tag, SpeciesName@, temp
StrArray SpeciesChangeTag
Array SpeciesChangeFlow
TagSelect SpeciesChanges
Checkbox SpeciesChangeOK@
Long SpciesChangeCount@, i
Real SpBalance@("Qm","t/h")
Sub Init()
SpeciesName = ClassTag()
EndSub
Sub Exec()
; find all SpeciesChanges
Criteria_Tag = Concatenate("([Links.Nett.Qm.", SpeciesName, " (g/h)]>0.001 OR [Links.Nett.Qm.", SpeciesName, " (g/h)]<-0.001)")
SpeciesChangeOK = SpeciesChanges.Exec(Criteria_Tag, false, false)
; find total number of SpeciesChanges
SpciesChangeCount = SpeciesChanges.GetLen()
; get all species change flows
flow_tag = Concatenate(".Links.Nett.Qm.",SpeciesName, " (t/h)")
SpeciesChanges.GetValues(flow_tag, SpeciesChangeFlow)
; sum all species change flows
SpBalance = SpeciesChangeFlow.Sum()
; get the unit list of where change occurred
SpeciesChangeTag.SetSize(SpciesChangeCount)
i=0
while (i<SpciesChangeCount)
temp = SpeciesChanges.Tag(i)
SpeciesChangeTag.SetAt(i, temp)
i = i + 1
endwhile
EndSub
EndClass
PageLabel "SpeciesBal"
TextLabel , "The species must be added to PlantModel-Settings Tab", ""
Class_SpeciesBalance # H2O, CuS
watch CuS.SpeciesChangeTag[all,8]
watch CuS.SpeciesChangeFlow[all,8]
Sub InitialiseSolution()
ForEachSub(Class_SpeciesBalance, Init())
CuS.SpeciesName = "CuS(s)"
EndSub
ForEachSub(Class_SpeciesBalance, Exec())
$
In the PGM file above, we have performed one calculations: Total CuS change, this includes input, output and any changes caused by reactions.
- This can be used to track the species movement across the process.
- This example is written as a class, other species can be easily added.
- These values will be shown on the General Controller Access Window when the solver stops.